Many life history strategies seem to be highly evolutionarily labile, evolving independently multiple times across a phylogeny. Dormancy seems to fit this bill, appearing to evolve rapidly within closely related clades. Our transcriptomic studies also support this view, showing that at the level of transcription, regulation of dormancy is surprisingly non-conserved in invertebrates, though different species seem to converge on a few similar physiological phenotypes. As we continue to accumulate transcriptomic data, we are further characterizing these metabolic and regulatory similarities to ask 1) is there any core cassette of pathways that are always associated with insect diapause, and 2) are pathways critical to diapause coopted from other highly conserved pathways such as stress responses.
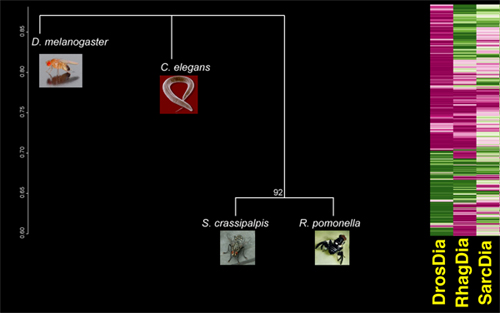
Clustering diagram based on the transcriptome-wide diapause response, and heat map of a subset of genes for insect representatives. Clustering illustrates widely divergent responses among insects, so much so that Drosophila diapause clusters no closer to other fly species than worm dormancy (C. elegans dauer stage). Yet, expression patterns illustrate that there are some clear commonalities in insects, particularly in genes linked to developmental arrest.
Though photoperiod may often regulate diapause, many species seem to have some sort of internal timer that dictates how long they stay in diapause. Recent studies from our group suggest that modulation of very slow, but not completely arrested rates of development during diapause regulate this process. We are currently using various developmental techniques to document this slow development and describe how it ultimately determines life history timing.
Confocal image of CNS Axon staining (green) against DAPI (blue) in the diapausing brain of Rhagoletis pomonella. Credit: Matin Sanaei and Jantina Toxopeus.